Sequence
Alignment
In bioinformatics,
a sequence alignment is a way of arranging the primary sequences of DNA, RNA, or
protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary
relationships between the sequences. Aligned sequences of nucleotide or amino
acid residues are typically
represented as rows within a matrix.
Gaps are inserted between the residues so that residues with identical or
similar characters are aligned in successive columns.
If two sequences in
an alignment share a common ancestor, mismatches
can be interpreted as point mutations and gaps as indels (that is, insertion or
deletion mutations) introduced in one or both lineages in the time since they diverged from one another. In
protein sequence alignment, the degree of similarity between amino acids
occupying a particular position in the sequence can be interpreted as a rough
measure of how conserved a
particular region or sequence motif is among lineages. The absence of
substitutions, or the presence of only very conservative substitutions (that
is, the substitution of amino acids whose side chains have similar biochemical
properties) in a particular region
of the sequence, suggest that this region has structural or functional
importance. Although DNA and RNA nucleotide bases are more similar to each
other than to amino acids, the conservation of base pairing can indicate a
similar functional or structural role. Sequence alignment can be used for
non-biological sequences, such as identifying similarities in a series of
letters and words present in human language.
Task 1: Vocabulary
– Match the words in the table below with their definitions
|
word |
meaning |
ans |
|
1 functional |
A that which is
left over |
1 |
|
2 structural |
B something
dissimilar or inappropriate |
2 |
|
3 evolutionary |
C having a high degree
of similarity in the primary or higher structures of homologous proteins |
3 |
|
4 residue |
D concerned with
the architecture and shape of proteins and nucleic acids |
4 |
|
5 matrix |
E area |
5 |
|
6 mismatch |
F the material or
tissue in which more specialized structures are embedded |
6 |
|
7 indel |
G related to what
something does rather than what it looks like |
7 |
|
8 lineage |
H a nucleotide or
amino acid sequence pattern that is widespread and seen as significant |
8 |
|
9 conserved |
I a ‘combined’
word relating to two types of genetic mutation |
9 |
|
10 sequence motif |
J change in
heritable traits determined by the shifts in the allele frequencies of genes |
10 |
|
11 region |
K a descending line
of offspring or an ascending line of parentage |
11 |
Task 2: Comprehension – Read the paragraph below and answer the following questions
Very short or very
similar sequences can be aligned by hand; however, most interesting problems require
the alignment of lengthy, highly variable or extremely numerous sequences that
cannot be aligned solely by human effort. Instead, human knowledge is primarily
applied in constructing algorithms to produce high-quality sequence alignments,
and occasionally in adjusting the final results to reflect patterns that are
difficult to represent algorithmically (especially in the case of nucleotide
sequences). Computational approaches to sequence alignment generally fall into
two categories: global alignments and local alignments. Calculating a global
alignment is a form of global optimization that "forces" the
alignment to span the entire length of all query sequences. By contrast, local
alignments identify regions of similarity within long sequences that are often
widely divergent overall. Local alignments are often preferable, but can be
more difficult to calculate because of the additional challenge of identifying
the regions of similarity. A variety of computational algorithms have been
applied to the sequence alignment problem, including slow but formally
optimizing methods like dynamic programming and efficient heuristic or
probabilistic methods designed for large-scale database search.
1
What kind of sequences can be aligned by hand?
2
What is the difference between global and local alignments?
3
Why are local alignments more difficult to calculate?
4
How is dynamic programming described?
Representations
Take the words listed below and insert them in the correct place
in the text.
colon, successive, graphically,
symbols, consensus, period,
nucleotide, conservativeness
Alignments are
commonly represented both __________ and in text format. In almost all sequence
alignment representations, sequences are written in rows arranged so that
aligned residues appear in __________ columns. In text formats, aligned columns
containing identical or similar characters are indicated with a system of
conservation__________. As in the image above, an asterisk or pipe symbol is
used to show identity between two columns; other less common symbols include a
__________ for conservative substitutions and a __________ for
semi-conservative substitutions. Many sequence visualization programs also use
color to display information about the properties of the individual sequence
elements; in DNA and RNA sequences, this equates to assigning each __________
its own color. In protein alignments, such as the one in the image above, color
is often used to indicate amino acid properties to aid in judging the __________
of a given amino acid substitution. For multiple sequences the last row in each
column is often the consensus sequence determined by the alignment.
Task 4: What is the following
paragraph about?
Sequence alignments
can be stored in a wide variety of text-based file formats, many of which were
originally developed in conjunction with a specific alignment program or
implementation. Most web-based tools allow a number of input and output
formats, such as FASTA format and GenBank format;
however, the use of specific tools authored by individual research laboratories
can be complicated by limited file format compatibility. A general conversion
program is available at READSEQ.
Global alignments,
which attempt to align every residue in every sequence, are most useful when
the sequences in the query set are similar and of roughly equal size. (This
does not mean global alignments cannot end in gaps.) A general global alignment
technique is called the Needleman-Wunsch algorithm
and is based on dynamic programming. Local alignments are more useful for
dissimilar sequences that are suspected to contain regions of similarity or
similar sequence motifs within their larger sequence context. The
Smith-Waterman algorithm is a general local alignment method also based on
dynamic programming. With sufficiently similar sequences, there is no
difference between local and global alignments.
Hybrid methods,
known as semiglobal or "glocal"
methods, attempt to find the best possible alignment that includes the start
and end of one or the other sequence. This can be especially useful when the
downstream part of one sequence overlaps with the upstream part of the other
sequence. In this case, neither global nor local alignment is entirely
appropriate: a global alignment would attempt to force the alignment to extend
beyond the region of overlap, while a local alignment might not fully cover the
region of overlap
Task 5: What would be a good
title for the above section of text?
Task 6: In the second paragraph
of this section two examples of ‘hybrids’ are given. What are they? How do you
think they got their names?
A The three primary
methods of producing pairwise alignments are
dot-matrix methods, dynamic programming, and word methods.
B Pairwise alignments can only be used between two sequences
at a time, but they are efficient to calculate and are often used for methods
that do not require extreme precision (such as searching a database for
sequences with high homology to a query).
C However, most
multiple sequence alignment techniques can align only two sequences. Although
each method has its individual strengths and weaknesses, all three methods have
difficulty with highly repetitive sequences of low information content -
especially where the number of repetitions differ in
the two sequences to be aligned.
D Pairwise sequence alignment methods are used to find the
best-matching piecewise (local) or global alignments of two query sequences.
Task 7: Jumbled text –
The section above has been jumbled. Put the sections in their
correct order to form a coherent paragraph.
|
1 |
clue |
2 |
clue |
3 |
clue |
4 |
clue |
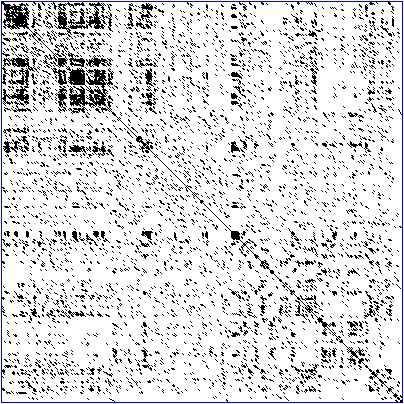
Dot-matrix
methods
The
dot-matrix approach, which implicitly produces a family of alignments for individual
sequence regions, is qualitative and simple,
though time-consuming to analyze on
a large scale. It is very easy to visually identify certain sequence
features—such as insertions, deletions, repeats, or inverted repeats—from a dot-matrix plot. To construct a dot-matrix
plot, the two sequences are written along the top row and leftmost column of a
two-dimensional matrix and a dot is placed at any point where the characters in
the appropriate columns match—this is a typical
recurrence plot. Some implementations vary the size or intensity of the dot
depending on the degree of similarity of the two characters, to accommodate conservative substitutions. The dot
plots of very closely related sequences will appear as a single line along the
matrix's main diagonal.
Dot
plots can also be used to assess repetitiveness
in a single sequence. A sequence can be plotted against itself and regions that
share significant similarities will appear as lines off the main diagonal. This
effect can occur when a protein consists of multiple similar structural domains.
Task
8: Vocabulary – Look at the following words. Find
them in the text and decide what kind of word they are and then tick the
correct box in the table below. Then try to fill in the remaining boxes with examples
of the same basic word in a different form.
|
Word in text |
noun |
verb |
adjective |
adverb |
more |
|
simple |
|
|
|
|
|
|
analyze |
|
|
|
|
|
|
inverted |
|
|
|
|
|
|
typical |
|
|
|
|
|
|
conservative |
|
|
|
|
|
|
repetitiveness |
|
|
|
|
|
|
structural |
|
|
|
|
|
Task 9a: Mini-presentation. Group A read
the section entitled Dynamic Programming and later explain it to the rest of
the class.
Dynamic programming
The technique of dynamic
programming can be applied to produce global alignments via the Needleman-Wunsch algorithm, and local alignments via the
Smith-Waterman algorithm. In typical usage, protein alignments use a
substitution matrix to assign scores to amino-acid matches or mismatches, and a
gap penalty for matching an amino acid in one sequence to a gap in the other.
DNA and RNA alignments may use a scoring matrix, but in practice often simply
assign a positive match score, a negative mismatch score, and a negative gap penalty.
(In standard dynamic programming, the score of each amino acid position is
independent of the identity of its neighbors, and therefore base stacking
effects are not taken into account. However, it is possible to account for such
effects by modifying the algorithm.)
Dynamic programming
can be useful in aligning nucleotide to protein sequences, a task complicated
by the need to take into account frameshift mutations
(usually insertions or deletions). The framesearch
method produces a series of global or local pairwise
alignments between a query nucleotide sequence and a search set of protein
sequences, or vice versa. Although the method is very slow, its ability to
evaluate frameshifts offset by an arbitrary number of
nucleotides makes the method useful for sequences containing large numbers of indels, which can be very difficult to align with more
efficient heuristic methods. In practice, the method requires large amounts of
computing power or a system whose architecture is specialized for dynamic programming.
The BLAST and EMBOSS suites provide basic tools for creating translated
alignments (though some of these approaches take advantage of side-effects of
sequence searching capabilities of the tools). More general methods are
available from both commercial sources, such as FrameSearch,
distributed as part of the Accelrys GCG package, and
Open Source software such as Genewise.
The dynamic
programming method is guaranteed to find an optimal alignment given a
particular scoring function; however, identifying a good scoring function is
often an empirical rather than a theoretical matter. Although dynamic
programming is extensible to more than two sequences, it is prohibitively slow
for large numbers of or extremely long sequences.
Task 9b: Mini-presentation. Group B read the section entitled Word
Methods and later explain it to the rest of the class.
Word methods
Word methods, also
known as k-tuple methods, are heuristic methods that
are not guaranteed to find an optimal alignment solution, but are significantly
more efficient than dynamic programming. These methods are especially useful in
large-scale database searches where it is understood that a large proportion of
the candidate sequences will have essentially no significant match with the
query sequence. Word methods are best known for their implementation in the
database search tools FASTA and the BLAST family. Word methods identify a
series of short, nonoverlapping subsequences
("words") in the query sequence that are then matched to candidate
database sequences. The relative positions of the word in the two sequences
being compared are subtracted to obtain an offset; this will indicate a region
of alignment if multiple distinct words produce the same offset. Only if this
region is detected do these methods apply more sensitive alignment criteria;
thus, many unnecessary comparisons with sequences of no appreciable similarity
are eliminated.
In the FASTA
method, the user defines a value k to use as the word length with which to
search the database. The method is slower but more sensitive at lower values of
k, which are also preferred for searches involving a very short query sequence.
The BLAST family of search methods provides a number of algorithms optimized
for particular types of queries, such as searching for distantly related
sequence matches. BLAST was developed to provide a faster alternative to FASTA
without sacrificing much accuracy; like FASTA, BLAST uses a word search of
length k, but evaluates only the most significant word matches, rather than
every word match as does FASTA. Most BLAST implementations use a fixed default
word length that is optimized for the query and database type, and that is
changed only under special circumstances, such as when searching with
repetitive or very short query sequences. Implementations can be found via a
number of web portals, such as EMBL FASTA and NCBI BLAST.
The IDF method
identifies weighted n-gram sequence fragments in large genomic databases whose indexing
characteristics permit the construction of indexed, sequence retrieval programs
where query processing time is determined mainly by the size of the query and
number of sequences retrieved rather than the number of sequences scanned. The
weighting scheme is based on the inverse document frequency (IDF) method, a
weighting formula that calculates the relative importance of indexing terms
based on term distribution. GPL open-source application
Task 9c: Mini-presentation. Group C read the sections entitled Multiple sequence alignments and Dynamic programming and
later explain them to the rest of the class.
Multiple sequence alignment
Multiple sequence
alignment (MSA) is an extension of pairwise alignment
to incorporate more than two sequences at a time. Multiple alignment methods
try to align all of the sequences in a given query set. Multiple alignments are
often used in identifying conserved sequence regions across a group of
sequences hypothesized to be evolutionarily related. Such conserved sequence
motifs can be used in conjunction with structural and mechanistic information
to locate the catalytic active sites of enzymes. Alignments are also used to
aid in establishing evolutionary relationships by constructing phylogenetic trees. MSAs are
computationally difficult to produce and most formulations of the problem lead
to NP-complete combinatorial optimization problems Nevertheless, the utility of
these alignments in bioinformatics has led to the development of a variety of
methods suitable for aligning three or more sequences.
Dynamic programming
The technique of
dynamic programming is theoretically applicable to any number of sequences;
however, because it is computationally expensive in both time and memory, it is
rarely used for more than three or four sequences in its most basic form. This
method requires constructing the n-dimensional equivalent of the sequence
matrix formed from two sequences, where n is the number of sequences in the
query. Standard dynamic programming is first used on all pairs of query
sequences and then the "alignment space" is filled in by considering
possible matches or gaps at intermediate positions, eventually constructing an
alignment essentially between each two-sequence alignment. Although this
technique is computationally expensive, its guarantee of a global optimum
solution is useful in cases where only a few sequences need to be aligned
accurately. One method for reducing the computational demands of dynamic
programming, which relies on the "sum of pairs" objective function,
has been implemented in the MSA software package
Task 9d: Mini-presentation. Group D read the sections entitled
Progressive methods, Iterative methods and Motif finding and later explain them
to the rest of the class.
Progressive methods
Progressive,
hierarchical, or tree methods generate an MSA by first aligning the most
similar sequences and then adding successively less related sequences or groups
to the alignment until the entire query set has been incorporated into the
solution. The initial tree describing the sequence relatedness is based on pairwise comparisons that may include heuristic pairwise alignment methods similar to FASTA. Progressive
alignment results are dependent on the choice of "most related"
sequences and thus can be sensitive to inaccuracies in the initial pairwise alignments. Most progressive MSA methods
additionally weight the sequences in the query set according to their
relatedness, which reduces the likelihood of making a poor choice of initial
sequences and thus improves alignment accuracy.
Many variations of
the Clustal progressive implementation are used for
multiple sequence alignment, phylogenetic tree
construction, and as input for protein structure prediction. A slower but more
accurate variant of the progressive method is known as T-Coffee.
Iterative methods
attempt to improve on the weak point of the progressive methods, the heavy
dependence on the accuracy of the initial pairwise
alignments. Iterative methods optimize an objective function based on a
selected alignment scoring method by assigning an initial global alignment and
then realigning sequence subsets. The realigned subsets are then themselves
aligned to produce the next iteration's MSA.
Motif finding, also
known as profile analysis, constructs global MSAs
that attempt to align short conserved sequence motifs among the sequences in
the query set. This is usually done by first constructing a general global MSA,
after which the highly conserved regions are isolated and used to construct a
set of profile matrices. The profile matrix for each conserved region is
arranged like a scoring matrix but its frequency counts for each amino acid or
nucleotide at each position are derived from the conserved region's character
distribution rather than from a more general empirical distribution. The
profile matrices are then used to search other sequences for occurrences of the
motif they characterize. In cases where the original data set contained a small
number of sequences, or only highly related sequences, pseudocounts
are added to normalize the character distributions represented in the motif.
Techniques inspired by
computer science
A variety of
general optimization algorithms commonly used in computer science have also been applied to the multiple sequence alignment
problem. Most recently hidden Markov models have been used to produce probability scores for a family of
possible MSAs for a given query set. Genetic algorithms
and simulated annealing have also been used in optimizing MSA scores as judged
by a scoring function like the sum-of-pairs method. More complete details and
software packages can be found in
the main article multiple sequence alignment.
Task 10: Active/Passive. Look at the two highlighted verbs in the
passage above. Are they active or passive form? Change them into the other
form.
Structural alignment
Structural
alignments, which are usually specific to protein and sometimes RNA sequences,
use information about the secondary and tertiary structure of the protein or
RNA molecule to aid in aligning the sequences. (1)These methods can be used for two or more sequences and typically
produce local alignments; however, because (2)they depend on the availability
of structural information, they can only be used for sequences whose
corresponding structures are known (usually through X-ray crystallography or
NMR spectroscopy). Because both protein and RNA structure is more
evolutionarily conserved than sequence, structural alignments can be more
reliable between sequences that are very distantly related and that have
diverged so extensively that sequence comparison cannot reliably detect (3)their
similarity.
Structural
alignments are used as the "gold standard" in evaluating alignments
for homology-based protein structure prediction because (4)they explicitly align regions of
the protein sequence that are structurally similar rather than relying
exclusively on sequence information. However, clearly structural alignments
cannot be used in structure prediction because at least one sequence in the
query set is the target to be modeled, for which the structure is not known.
(5)It has been shown that, given the
structural alignment between a target and a template sequence, highly accurate
models of the target protein sequence can be produced; a major stumbling block
in homology-based structure prediction is the production of structurally
accurate alignments given only sequence information.
Task 11: Pronoun Referents. Look at the
highlighted words in the passage above. What do they refer to?
|
Word |
Refers to… |
|
1 these |
|
|
2 they |
|
|
3 their |
|
|
4 they |
|
|
5 it |
|
DALI
The DALI method,
or distance matrix alignment, is a fragment-based method for constructing
structural alignments based on contact similarity patterns between successive hexapeptides in the query sequences. It can generate pairwise or multiple alignments and identify a query
sequence's structural neighbors in the Protein Data Bank (PDB). It has been
used to construct the FSSP structural alignment database (Fold
classification based on Structure-Structure alignment of Proteins, or Families
of Structurally Similar Proteins). A DALI webserver
can be accessed at EBI DALI and the FSSP is located at The Dali Database.
SSAP
SSAP (sequential structure alignment program) is
a dynamic programming-based method of structural alignment that uses
atom-to-atom vectors in structure space as comparison points. It has been
extended since its original description to include multiple as well as pairwise alignment, and has been used in the construction
of the CATH
(Class, Architecture, Topology, Homology) hierarchical database classification
of protein folds The CATH database can be accessed at CATH Protein Structure
Classification.
The combinatorial
extension (CE)
method of structural alignment generates a pairwise
structural alignment by using local geometry to align short fragments of the
two proteins being analyzed and then assembles these fragments into a larger
alignment. Based on measures such as rigid-body root mean square distance, residue
distances, local secondary structure, and surrounding environmental features
such as residue neighbor hydrophobicity, local
alignments called "aligned fragment pairs" (AFPs)
are generated and used to build a similarity matrix representing all possible
structural alignments within predefined cutoff criteria. A path from one
protein structure state to the other is then traced through the matrix by
extending the growing alignment one fragment at a time. The optimal such path
defines the CE alignment. A web-based server implementing the method and
providing a database of pairwise alignments of
structures in the PDB is located at the Combinatorial Extension website.
Task 12: Abbreviations. Look at the highlighted abbreviations in
the passage above. What do they stand for?
|
Abbreviation |
Means… |
|
DALI |
|
|
FSSP |
|
|
SSAP |
|
|
CATH |
|
|
CE |
|
|
AFP |
|
Phylogenetics and sequence alignment are closely related fields
due to the shared necessity of evaluating sequence relatedness. The field of phylogenetics makes extensive use of sequence alignments in
the construction and interpretation of phylogenetic
trees, which are used to classify the evolutionary relationships between
homologous genes represented in the genomes of divergent species. The degree to
which sequences in a query set differ is qualitatively related to the
sequences' evolutionary distance from one another. Roughly speaking, high
sequence homology suggests that the sequences in question have a comparatively
young most recent common ancestor, while low homology suggests that the
divergence is more ancient. This approximation, which reflects the
"molecular clock" hypothesis that a roughly constant rate of
evolutionary change can be used to extrapolate the elapsed time since two genes
first diverged (that is, the coalescence time), assumes that the effects of
mutation and selection are constant across sequence lineages. Therefore it does
not account for possible difference among organisms or species in the rates of
DNA repair or the possible functional conservation of specific regions in a
sequence. (In the case of nucleotide sequences, the molecular clock hypothesis
in its most basic form also discounts the difference in acceptance rates
between silent mutations that do not alter the meaning of a given codon and other mutations that result in a different amino
acid being incorporated into the protein.) More statistically accurate methods
allow the evolutionary rate on each branch of the phylogenetic
tree to vary, thus producing better estimates of coalescence times for genes.
Progressive
multiple alignment techniques produce a phylogenetic
tree by necessity because they incorporate sequences into the growing alignment
in order of relatedness. Other techniques that assemble MSAs
and phylogenetic trees score and sort trees first and
calculate an MSA from the highest-scoring tree. Commonly used methods of phylogenetic tree construction are mainly heuristic because
the problem of selecting the optimal tree, like the problem of selecting the
optimal MSA, is NP-hard
Task 13: Comprehension. Read the passage above and answer the
following questions
1 What are phylogenic trees used for?
2 In what way are high sequence homology and
low homology different?
3 What is coalescence time?
4 Why are the most frequently
used ways of building a phylogenic tree mainly heuristic?
Homework Tasks: Make up 5
vocabulary matching, 2 pronuoun referent questions
and 5 comprehension questions from the Assessment of significance, scoring functions and software sections below.
Sequence alignments
are useful in bioinformatics for identifying sequence similarity, producing phylogenetic trees, and developing homology models of
protein structures. However, the biological relevance of sequence alignments is
not always clear. Alignments are often assumed to reflect a degree of
evolutionary change between sequences descended from a common ancestor;
however, it is formally possible that convergent evolution can occur to produce
apparent similarity between proteins that are evolutionarily unrelated but
perform similar functions and have similar structures.
In database
searches such as BLAST, statistical methods can determine the likelihood of a
particular alignment between sequences or sequence regions arising by chance
given the size and composition of the database being searched. These values can
vary significantly depending on the search space. In particular, the likelihood
of finding a given alignment by chance increases if the database consists only
of sequences from the same organism as the query sequence. Repetitive sequences
in the database or query can also distort both the search results and the
assessment of statistical significance; BLAST automatically filters such
repetitive sequences in the query to avoid apparent hits that are statistical
artifacts.
The choice of a
scoring function that reflects biological or statistical observations about known
sequences is important to producing good alignments. Protein sequences are
frequently aligned using substitution matrices that reflect the probabilities
of given character-to-character substitutions. A series of matrices called PAM
matrices (Percent Accepted Mutation matrices, originally defined by Margaret Dayhoff and sometimes referred to as "Dayhoff matrices") explicitly encode evolutionary
approximations regarding the rates and probabilities of particular amino acid
mutations. Another common series of scoring matrices, known as BLOSUM (Blocks
Substitution Matrix), encodes empirically derived substitution probabilities.
Variants of both types of matrices are used to detect sequences with differing
levels of divergence, thus allowing users of BLAST or FASTA to restrict
searches to more closely related matches or expand to detect more divergent
sequences. Gap penalties account for the introduction of a gap - on the
evolutionary model, an insertion or deletion mutation - in both nucleotide and
protein sequences, and therefore the penalty values should be proportional to
the expected rate of such mutations. The quality of the alignments produced
therefore depends on the quality of the scoring function.
It can be very
useful and instructive to try the same alignment several times with different
choices for scoring matrix and/or gap penalty values and compare the results.
By noting which regions look similar no matter what settings are used, and
which look different, one can get an excellent sense
of where the algorithm had difficulty finding a robust solution.
Common software
tools used for general sequence alignment tasks include ClustalW
and T-coffee for alignment, and BLAST for database
searching.
Alignment
algorithms and software can be directly compared to one another using a
standardized set of benchmark reference multiple sequence alignments known as BAliBASE. The dataset consists of structural alignments,
which can be considered a standard against which purely sequence-based methods
are compared. The relative performance of many common alignment methods on
frequently encountered alignment problems has been tabulated and selected
results published online at BAliBASE
Task 14: Image indentification. Look at the three
captions below and link them to the following images. Then decide where in the
text they should have appeared.
1
Dot plot of a human zinc-finger transcription factor (GenBank
NM_002383) against itself to show self-similarity
2 A sequence
alignment, produced by ClustalW between two human
zinc finger proteins identified by Gen Bank accession number
3
Illustration of global and local alignments demonstrating the 'gappy' quality of global alignments that can occur if
sequences are insufficiently similar
Image A

Image B

Image C