Human
Genome: in search of normality
In this
unit are going to read an article about the human genome
Before
reading
Task 1
Imagine you are working on compiling a dictionary.With a partner
prepare and write down a definition of the human genome.
Task
2
Make a list of 10 nouns you might expect to find in an article about
the human genome
Task
3
Make a list of 10 verbs you might find in an article about the human
genome
Task
4
With a partner discuss why research into the human genome can be
beneficial to society.
While
reading
Task 1
Read paragraph 1 and, with information found in the text, fill in the
chart below.
Chart 1
News and Views
Nature
444, 428-429 (23 November 2006) | doi:10.1038/444428a; Published online
22 November 2006
Human genomics: In search of
normality
Kevin V. Shianna1 and
Huntington F. Willard1
The first map of copy-number variation
in the human genome has been created. It is now feasible to examine the
role of such genome variation in disease and to explore in depth the
extent of 'normal' variability.
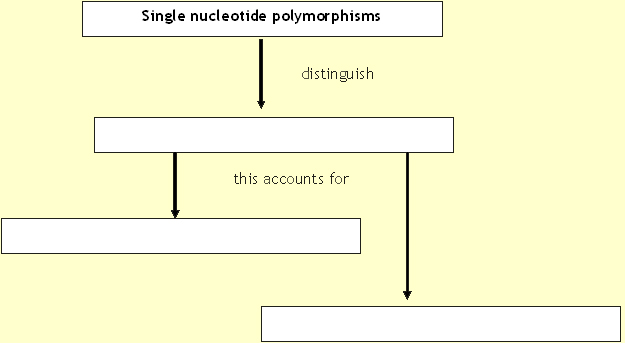
The human genome contains many
forms of genetic variation. The most plentiful are the millions of
single base-pair changes in the DNA code that were identified in the
course of determining the human genome sequence, and then more
systematically through the International HapMap Project1. These
so-called single nucleotide polymorphisms (SNPs) distinguish any two
unrelated copies of the genome. They account for the long-hypothesized,
evolutionarily 'neutral' forms of widespread genetic variation that
mark diversity within our species, as well as mutations, both rare and
common, that account for or contribute to disease.
Task
2
Read paragraph 2 and, with information found in the text, fill in the
chart 2 and 3 below.
Less expected have been
variations in the copy number of sequence elements — that is, variation
in the number of deleted or duplicated versions of segments of the
genome that result in a range of the number of copies (instead of the
usual two) among apparently 'normal' members of the population2.
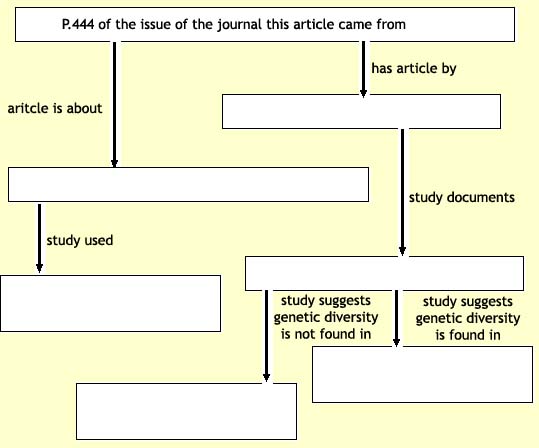
Several studies have described the prevalence of common deletion
polymorphisms in the human genome3, 4. On page 444 of this issue, Redon
et al.5 now present results of a global genome-wide screen looking for
all types of copy-number variants (CNVs) using several hundred
reference samples from four human populations. They document nearly
1,500 variable regions, covering a remarkable 12% of the human genome
and including hundreds of genes and other functional elements whose
copy number differs, sometimes dramatically, among us. The data suggest
that the greatest source of genetic diversity in our species lies not
in millions of SNPs, but rather in larger segments of the genome whose
presence or absence calls into question what exactly is a 'normal'
human genome.
Chart 2
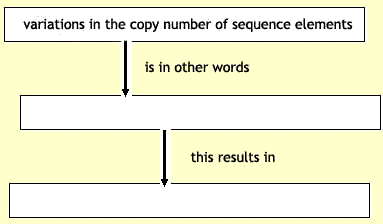
Chart 3
Task 3
Read paragraph 3 and in your own words define -
1.genotype
2. allele
3. segment
To detect CNVs,
Redon et al.
used two complementary genome-wide technologies. The first was a
genotyping approach in which some 500,000 SNPs were assayed, looking
for stretches of adjacent SNPs that displayed atypical ratios of the
expected two versions (called alleles) of a given SNP. The second
involved comparing each sample with a reference standard, and looking
for systematic differences in intensity among a set of more than 26,000
large-insert cloned segments that span nearly all of the currently
sequenced portion of the genome.
Task
4
Read
paragraph 4 and complete the following sentence -
_______________________ was between fifty and one hundred times less
than_____________________.
Combining these
approaches
provided coverage adequate to detect most forms of CNVs. In total,
1,447 CNVs were identified across the 270 HapMap samples. The estimated
average length of CNV regions per genome analysed was more than 20
million base pairs, representing some 5- to 10-fold more variation
between any two randomly chosen genomes than suggested previously by
studying SNPs alone. More than half of the CNVs that were identified
overlap known annotated genes in the genome. So it is likely that CNVs
play a role in so-called complex diseases, in which multiple genes
and/or gene–environment interactions are involved.
Task 5
Read
paragraph 5 and answer the following questions.
1. What does 'classically' mean?
2. What is described as being 'classical' in this paragraph?
3. Do scientists still believe this? If the answer is 'yes', why? If
the answer is 'no' than why not?
Mechanistically, how
might
copy-number variation be involved with complex disease? When deletions
or duplications are present within a gene or its regulatory region,
there is a reasonable chance that there will be an imbalance in the
appropriate level of RNA and thus protein production from that gene.
For genes and pathways in which the amount of a functional product
produced is critical, it seems likely that CNVs could underscore
variation in susceptibility to disease. Classically, variation in the
copy number of the globin genes was shown to be responsible for various
disorders of haemoglobin, such as the -thalassaemias6. More
recently, variable copy number of the CCL3L1 gene was reported to be
associated with increased resistance to infection by HIV7.
Task
6
Read
paragraph 6. What do the highlighted and underlined words refer to -
Many genome-wide studies are
currently under way that aim to find SNPs associated with complex
disease. These
studies in effect look for disease- and
population-specific changes in the frequencies of SNP alleles, using
arrays containing 'tagging SNPs' that act as proxies for other closely
associated SNPs that are inherited together as a block. The likely
involvement of CNVs in complex disease, however, raises the question of
whether the many CNVs reported by Redon et al.5 can also be detected by
association with one of these tagging SNPs. The answer seems to be both
yes and no. Some CNVs may be associated with their
neighbouring SNPs
over time, but others may be of newer origin and their
presence or
absence may not be accurately tagged. Thus, densely spaced SNPs will
probably be a prerequisite for the most efficient use of CNVs in
genome-wide SNP-based association studies.
Word
|
refers
to...
|
| These |
|
| their |
|
| their |
|
Task
7
Read
paragraph 7 and think of synonyms (words or phrases) for the
highlighted and underlined terms.
Given the limited set of
reference samples assayed, the 1,500 CNVs reported by Redon et al. are
probably the tip
of the
iceberg. As the results and the raw
data
from
the first wave of genome-wide association studies become available, it
will be essential to catalogue the full range of human CNVs. A complete
map of CNVs in global populations will be necessary before we can fully
understand which of the variants have clinical or other consequences,
and which are, in fact, within the extremes of what we consider
'normal'.
Word
or phrase
|
Synonym
|
| tip
of the
iceberg |
|
| raw
data |
|
| consequences |
|
Task
8
Read
paragraph 8 and put the following events in chronological order.
1. Mendel laws of inheritance were discovered again.
2. Scientists became aware of DNA.
3. Archibald Garrod refer to biochemical variants as "chemical
indiviuality".
More than a hundred
years ago
— even before the rediscovery of Mendel's laws of inheritance, and well
before an awareness of DNA and genomes — the physician Sir Archibald
Garrod first shed light on what he termed "chemical individuality" to
refer to biochemical variants that shape the intricacies of metabolism
in different individuals8. As he observed9 so presciently, "the
existence of chemical individuality follows of necessity from that of
chemical specificity, but we should expect the differences between
individuals to be still more subtle and difficult of detection." Our
current view of "genomic individuality" at the level of SNPs, CNVs and
chromosomal variants indeed extends his view in ways "more subtle and
difficult of detection". The stage is set for global studies to explore
anew, as Garrod once did, the clinical significance of human variation.
First
event
|
|
Second
event
|
|
Third
event
|
|
Task 9
Read
the references and find when A.E. Garrod's paper on Molecular Medicine
was first published.
References
1.
International HapMap Consortium Nature 437, 1299–1320 (2005).
2. Feuk, L.
et al. Nature Rev. Genet. 7, 85–97 (2006).
3. Hinds, D.
A. et al. Nature Genet. 38, 82–85 (2005).
4.
McCarroll, S. A. et al. Nature Genet. 38, 86–92 (2005).
5. Redon, R.
et al. Nature 444, 444–454 (2006).
6.
Weatherall, D. J. Am. J. Hum. Genet. 74, 385–392 (2004).
7. Gonzalez,
E. et al. Science 307, 1434–1440 (2005).
8. Garrod,
A. E. Mol. Med. 2, 274–282 (1996; reprint of original 1902 paper).
9. Garrod,
A. Inborn Errors of Metabolism 2nd edn (Oxford Univ. Press, 1923).



